
(System Python)
In an earlier work a SoC system using the AVR processor core has
been developed using SysPy, for the implementation of the Sobel algorithm
for image processing. Using the Leon core and a custom
co-processor another complete SoC system has been designed. The
custom core has been designed using VHDL and added to SysPyís
component libraries. The core performs simulations of biochemical
reaction networks (BioModels) using Gillespieís Stochastic Simulator
Algorithm (SSA). The core can be used to simulate in hardware
BioModels with thousands of reactions as needed in large scale
system biology studies.
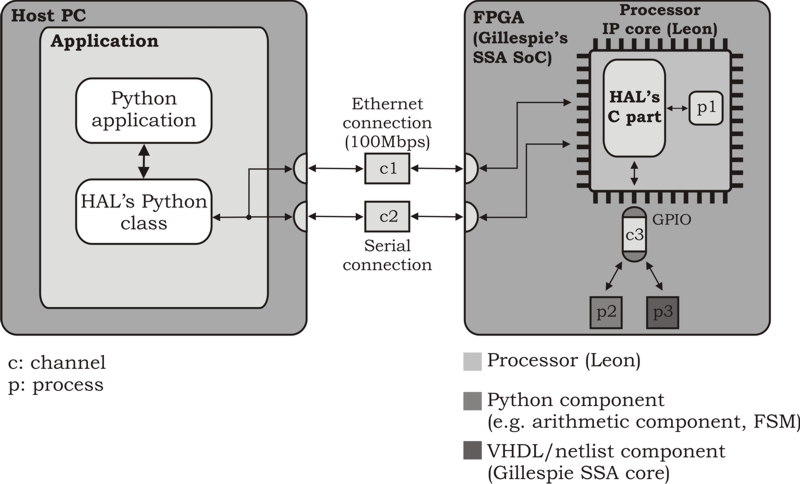
Since we can easily design processor-centric systems using
SysPy, we decided also to develop a Hardware Abstraction Layer
(HAL) as a Python interface software, that gives to the user access
to the available resources on the FPGA and the Leon processor. The
HALís software is divided into two parts: a) a Python part running
on the host PC and b) a part developed in C running on Leon. The
part on the PC is a Python class with various functions which can be
used to access the ethernet, the serial and the GPIO connection of
the processor.
A Python application can be developed that will create a HAL
object and access all its available functions. Ethernet and GPIO
functions use the processorís RAM memory as a buffer during data
transmissions. Using HAL, different processes can be initiated, running
either on the processor or on custom components connected to
the processorís GPIO ports.
The HALís functions have been tested on Gillespieís SoC. The
application, through HALís functions, uses the serial connection as a
control channel and the ethernet and GPIO connections as data channels.
Leonís RAM DDR2 memory is used as a data buffer, while the
algorithmís results are transmitted back to the PC through the ethernet
connection and stored in Python arrays for further processing at
the host.
Furthermore upon selecting a BioModel, its SBML representation
can automatically be parsed to extract a number of parameters
of the Gillespie algorithm, e.g. the version of the algorithm to be
used, the number of reactions and chemical species in the model etc.
All these automatically extracted parameters are used to initialize the
SoCs Block RAMs.
After SoCs parametrization, a Tcl script creates a complete design
project for Xilinx tools and executes the synthesis and place and
route processes to generate the configuration bitstream file for FPGA
programming. The user of the system (presumably a life science expert
with no knowledge of FGPA programming) can rely on SysPy to
deliver the best SoC setup for his/her specific BioModel simulation
(based on the parameters) and deliver to the PC the simulation results
in an effortless yet much faster manner (relatively to a software
PC-based simulation).
The following figure shows HAL software interfacing a typical processor-centric SoC
configuration of the Leon processor and the Gillespie SSA component.
All the top-level Python files describing the Gillespie SoC, as well as
the generated VHDL files and the Tcl script used for the FPGA implemenetation in a Xilinx
Virtex-5 device, can be found below:
Python - VHDL descriptions
Main Page


